Revolution in Antibiotics: Pioneering a New Era of Antimicrobial Strategies
Other Articles

Disrupting Bacterial Transcription to Tackle Multidrug Resistance
Study conducted by Prof. Cong MAand his research team

The rapid emergence of antibiotic-resistant bacterial infections represents one of the most pressing challenges to global public health in the 21st century. Since the discovery of penicillin in the early 20th century, antibiotics have revolutionised medicine, saving countless lives by effectively treating bacterial infections. However, the overuse and misuse of these life-saving drugs have led to the evolution of multidrug-resistant pathogens, rendering many antibiotics ineffective. This has created an urgent need for innovative approaches to antibiotic discovery. One promising approach is targeting bacterial transcription, a fundamental and essential process for bacterial survival.
Transcription is the process by which bacteria convert their genetic information (DNA) into RNANote1, which is then used to produce proteins necessary for growth and function. It is carried out by the enzyme RNA polymerase (RNAP) and regulated by a series of protein transcription factors. RNAP and its associated transcription factors are highly conserved across the bacterial domain, making them attractive targets for broad-spectrum antibiotics (Figure 1a).
A research team led by Prof. Cong MA, Associate Professor of the Department of Applied Biology and Chemical Technology, has embarked on first-in-class antimicrobial drug discovery with unique mechanisms of action to tackle antimicrobial resistance. Their work focuses on disrupting protein−protein interactions (PPIs) within the bacterial transcription complex, offering a promising avenue for the development of new antimicrobial agents (Figure 1b) [1]. Typical single-target antibiotics are susceptible to antimicrobial resistance (AMR) due to resistance mutations on their targets, while protein−protein interaction requires complementary mutations to develop AMR and, meanwhile, to maintain robust interactions between them. During their research, the team has identified interactions between bacterial transcription factors NusB and NusE [2], as well as between bacterial RNA polymerase and the sigma factor [3], as promising drug targets. Recently, they identified the interaction between bacterial RNA polymerase and NusG as a new target for drug discovery, leading to a novel series of antimicrobial agents [4].
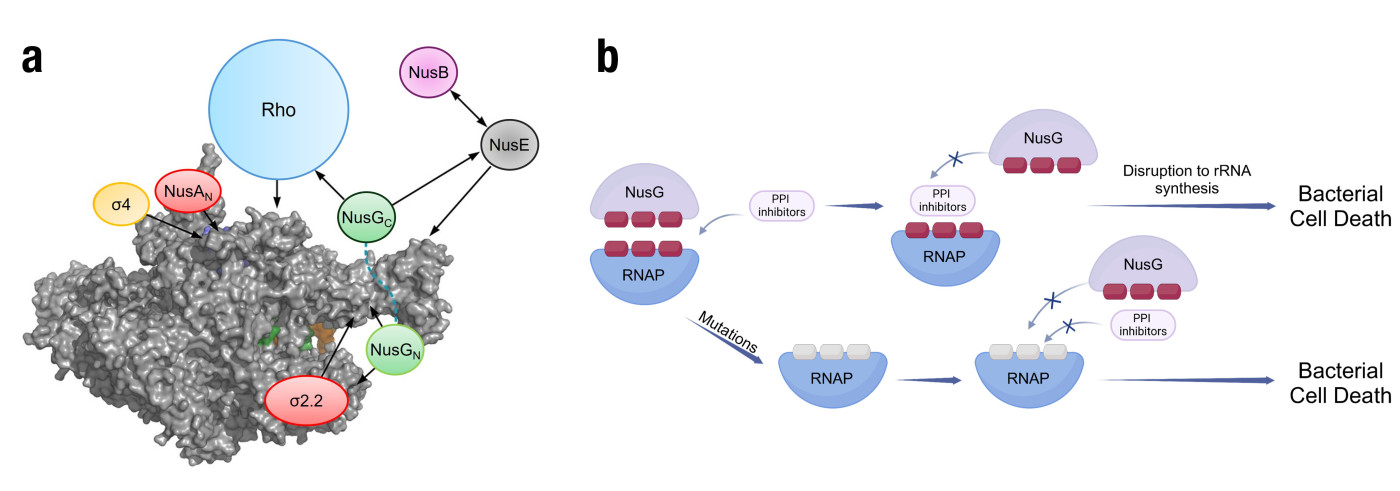
Figure 1. (a) Transcription factor interaction network. The space-filled RNAP elongation complex is shown in gray. Arrows indicate known interactions. All of these interactions could potentially be exploited in the development of new antimicrobial compounds. (b) Advantages of NusG-RNAP PPI inhibitors in combating AMR over typical antibiotics
Based on the crystal structure of E. coli RNAP bound to NusG (obtained from the Protein Data Bank database), key amino acids in the clamp-helix (CH) domain of RNAP that interact with NusG have been identified. These amino acids, particularly R270, R278, and R281, were used to generate the pharmacophore model (Figure 2). The screening was performed on the MiniMaybridge database, resulting in 57 possible compounds. Further evaluation through antibiotic susceptibility testing led to the discovery of a promising hit compound named AW00783. Through systematic modifications, the team synthesised derivatives with significantly improved potency. The most promising compound demonstrated notable activity against both Gram-positive and Gram-negative bacteria, with minimum inhibitory concentration (MIC) as low as 1 µg/mL against S. pneumoniae.
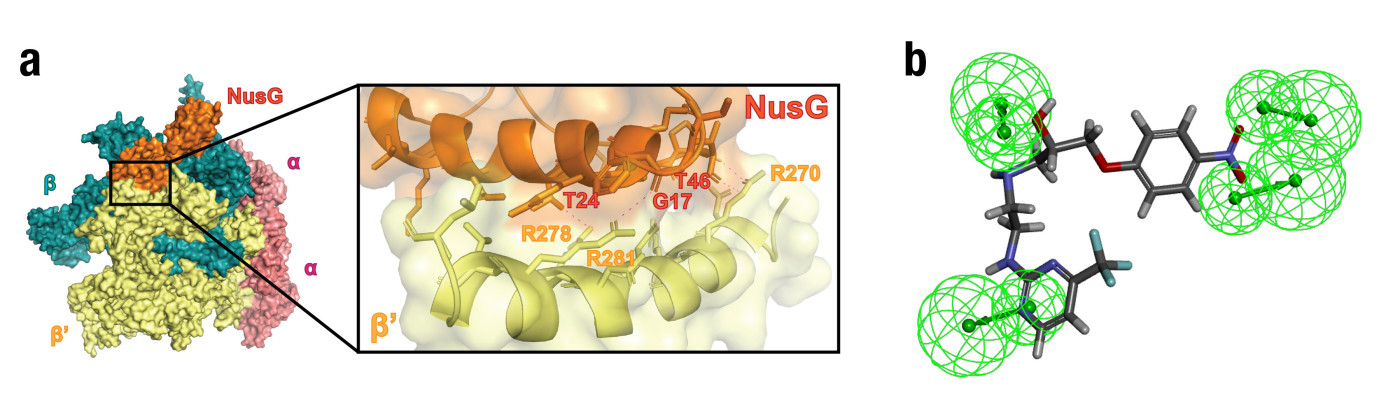
Figure 2. (a) Crystal structure of E. coli RNA polymerase core enzyme (subunits ααββ’) bound to NusG (PDB: 5TBZ). Left: the overall structure of a bacterial transcription complex, highlighting the key components of the core enzyme (Yellow: RNAP β’ subunit; Orange: NusG). Right: the key hydrogen bond interactions between NusG and the RNAP β’CH region. (b) Hit compound AW00783 docking to the pharmacophore model (Green spheres: hydrogen bond donor)
Key pharmacological assays were employed to validate the efficacy of these novel antibiotic candidates (Figure 3). The time-kill kinetics revealed that the antimicrobial agent gradually eradicated bacteria at a concentration of 1× MIC or higher, showing a bactericidal character. The protein−protein inhibition assay and fluorescent microscopy images confirmed that the antimicrobial agent inhibited with NusG and RNAP β’CH interaction and disrupted the subcellular localisation of the transcription complex, and an in-cell transcription assay further validated its mechanism of action. Together, these results provide robust evidence for the mechanism and potential of these compounds to combat resistant pathogens.
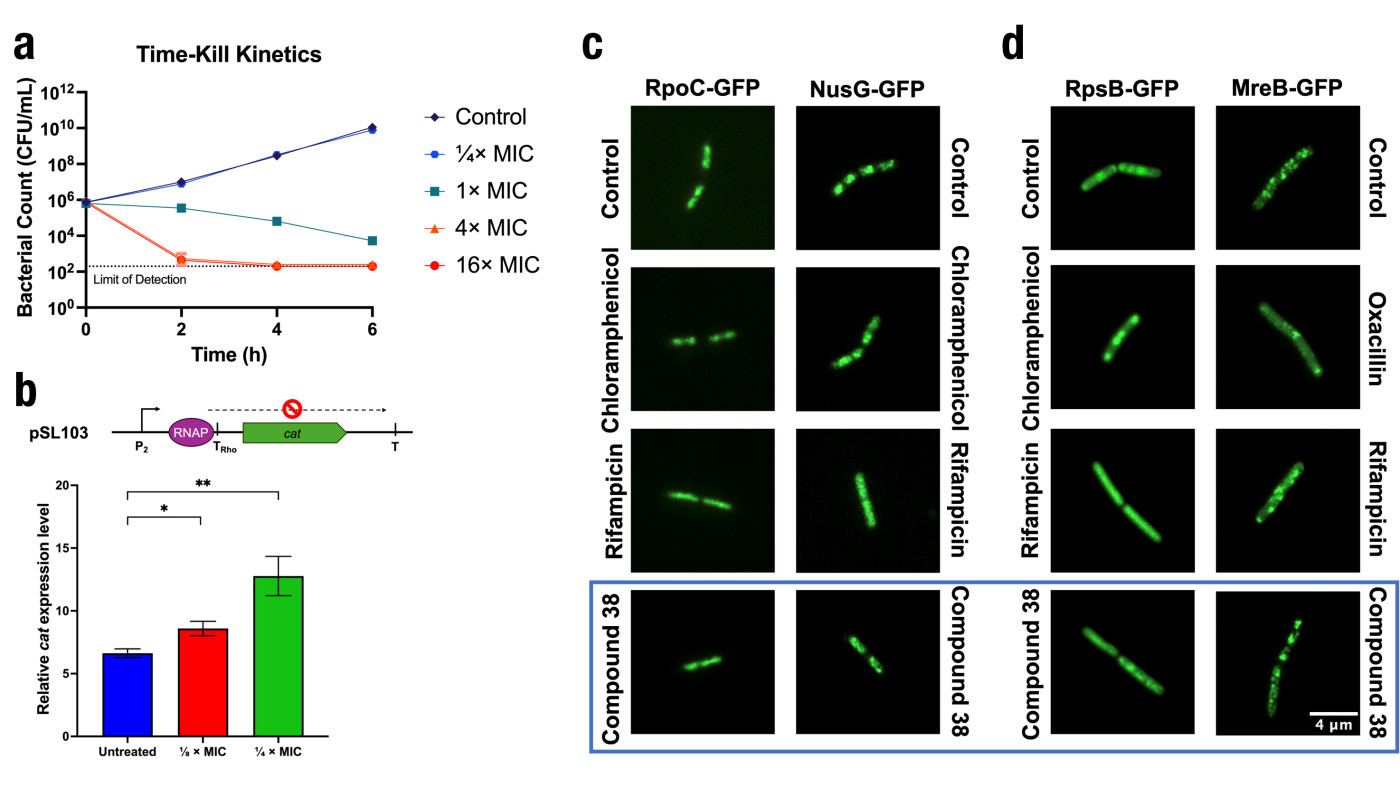
Figure 3. (a) Effect of compound 38 on the time-kill kinetics of CA-MRSA strain USA300 at 1/4×, 1×, 4× and 16×MIC in cation-adjusted Mueller-Hinton broth (CA-MHB) media. Experiments were performed in triplicates. (b) Cell-based transcription assay that showed compound effects against NusG function. The scheme of the plasmid pSL103 used was shown. Relative cat expression levels in pSL103 transformants treated differently. Technical repeats were performed to ensure data reproducibility. Statistical significance between treatment groups and untreated control was assessed using unpaired t-tests, with p < 0.05 denoted by * and p < 0.01 denoted by **. (c) Epifluorescence microscopy of B. subtilis with GFP tagged on transcription-related proteins. (d) Epifluorescence microscopy of B. subtilis with GFP tagged on non-transcription-related proteins. RpoC: RNAP β’CH, RpsB: ribosome RpsB, MreB: cell membrane-coordination MreB. The untreated control demonstrates no significant alteration in localisation, whereas the antibiotics-treated cells show a distinct pattern of delocalisation. Upon treatment with 1× MIC of compound 38 (fourth panel), cells exhibit a delocalisation, with the morphology resembling that observed in rifampicin-treated cells (third panel). Overall, compound 38 showed a similar bacterial cytological profile to rifampicin. Scale bar: 4 μm.
In silico docking studies were performed with Autodock Vina to simulate the docking model of candidate compound 38 with the β’CH of RNAP. The binding site was defined based on key residues, namely Arg270, Arg278, and Arg281, which were known to establish essential interactions with NusG. To evaluate the drug-like properties of synthesised compounds, absorption, distribution, metabolism and excretion properties (ADME) of the most potent compound 38 was predicted using Schrödinger Maestro 13.5 (Figure 4b). It was suggested that the selected compound possessed outstanding oral absorption, and low possibility of heart toxicity.
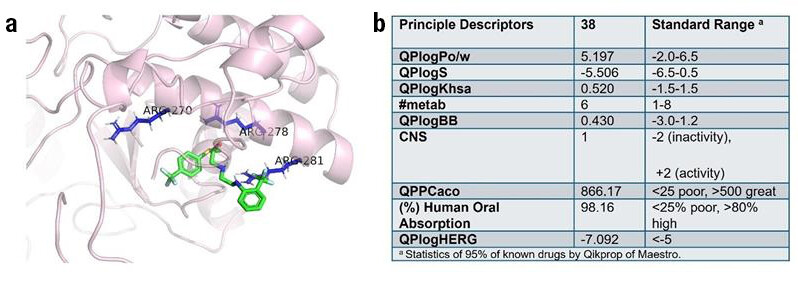
Figure 4. (a) Docking models of compound 38 with β’CH of RNAP. The ligand was coloured in green and the key residues (Arg270, Arg278, and Arg281) were coloured in blue. (b) In silico ADME evaluation of compound 38. Various pharmacokinetic and pharmacodynamic parameters were calculated: octanol−water partitioning coefficient (QPlogPo/w), aqueous solubility (QPlogS), binding to human serum albumin (QPlogKhsa), number of likely metabolic reactions (#metab), brain/blood partition coefficient (QPlogBB), central nervous system activity (CNS), apparent Caco-2 cell permeability (QPPCaco), human oral absorption, and predicted IC50 value for blockage of HERG K+ channels (QPloghERG).
Overall, this is the first reported research to explore the RNAP-NusG interaction in bacteria as a target for creating antimicrobial PPI inhibitors. A rational drug discovery strategy was employed to generate small-molecule antibiotics that target the β’CH of RNAP. These candidate compounds exhibited promising antimicrobial activities against both Gram-positive and Gram-negative pathogens. This research forms the basis for the structural optimisation of the lead compound to explore its potential as a new antimicrobial molecule targeting bacterial transcription.
The research is partly supported by the Hong Kong Research Grants Council (PolyU 15100021 and C5008-19G) and The Hong Kong Polytechnic University (1-ZE2E and State Key Laboratory of Chemical Biology and Drug Discovery). Supporting information is available free of charge at:
https://pubs.acs.org/doi/10.1021/acs.jmedchem.4c01386.
Prof. Ma is a co-founder of Ynno Med Ltd., a PolyU start-up developing new antibiotic drug and the award winner of the Falling Walls Science Breakthroughs of the Year 2023 under the category of Science Start-ups (Falling Walls Venture). Prof. Ma is also honoured the Global Innovation Award at TechConnect World Innovation Conference and Expo 2019 staged in the United States. Prof. Ma is currently the Director of PolyU Marshall Research Centre for Medical Microbial Biotechnology.
| Notes |
|---|
RNA (ribonucleic acid) is a nucleic acid found in all living cells. It resembles DNA structurally but it is typically single-stranded. Unlike DNA, which consists of deoxyribose, RNA’s backbone consists of alternating phosphate groups and ribose sugar.
| References |
|---|
[1] Ma, C., Yang, X., Lewis, P. J. (2016). Bacterial Transcription as a Target for Antibacterial Drug Development. Microbiology and Molecular Biology Reviews, 80 (1), 139-160. DOI: doi:10.1128/mmbr.00055-15
[2] Qiu, Y., Chan, S. T., Lin, L., Shek, T. L., Tsang, T. F., Barua, N., Zhang, Y., Ip, M., Chan, P. K.-s., Blanchard, N. (2019). Design, synthesis and biological evaluation of antimicrobial diarylimine and–amine compounds targeting the interaction between the bacterial NusB and NusE proteins. European journal of medicinal chemistry, 178, 214-231
[3] Ye, J., Chu, A. J., Harper, R., Chan, S. T., Shek, T. L., Zhang, Y., Ip, M., Sambir, M., Artsimovitch, I., Zuo, Z. (2020). Discovery of antibacterials that inhibit bacterial RNA polymerase interactions with sigma factors. Journal of medicinal chemistry, 63 (14), 7695-7720
[4] Zheng, Y., Kan, C. H., Tsang, T. F., Liu, Y., Liu, T., Tsang, M. W., Lam, L. Y., Yang, X., Ma, C. (2024). Discovery of Inhibitors Targeting Protein–Protein Interaction between Bacterial RNA Polymerase and NusG as Novel Antimicrobials. Journal of Medicinal Chemistry, 67 (18), 16556-16575
 | Prof. Cong MA |



