Quantum Leap in Molecular Vibronic Spectroscopy: Integrated Photonic Networks with Squeezed Vacuum States
Other Articles

A Scalable Quantum Photonic Microprocessor Enables the Simulation of Complex Molecular Vibronic Spectra
Study conducted by Prof. Lip Ket CHINand the research team led by Prof. Ai-Qun LIU

Understanding and predicting molecular vibronic spectra is a cornerstone of quantum technology, chemical analysis and biological labelling. These spectra, which arise from transitions involving both electronic and vibrational energy levels in molecules, are crucial for applications ranging from drug discovery to materials design. Yet, the computational challenge of generating accurate vibronic spectra for molecules with many vibrational modes is formidable.
Classical algorithms, such as the eigenvalue-trace method, scale exponentially with system size, making them impractical for large molecules. Even quantum-inspired classical approaches offer only partial solutions. While quantum simulation holds the promise of overcoming these barriers, current hardware faces limitations. In particular, superconducting qubits and trapped ions struggle with scalability, gate fidelity and operational temperature. These challenges restrict experimental demonstrations to molecules with just two vibronic modes. Photonic quantum systems, with their room-temperature operation and high controllability, offer a compelling alternative but practical, scalable implementations for real molecular systems have remained elusive.
In recent research published in Nature Communications [1], Prof. Lip Ket CHIN, Associate Professor of the Department of Electrical and Electronic Engineering at The Hong Kong Polytechnic University, and the research team led by Prof. Ai-Qun LIU from the same department address these challenges by introducing a novel algorithm and an integrated quantum photonic microprocessor chip capable of simulating molecular vibronic spectra with unprecedented scalability and fidelity.
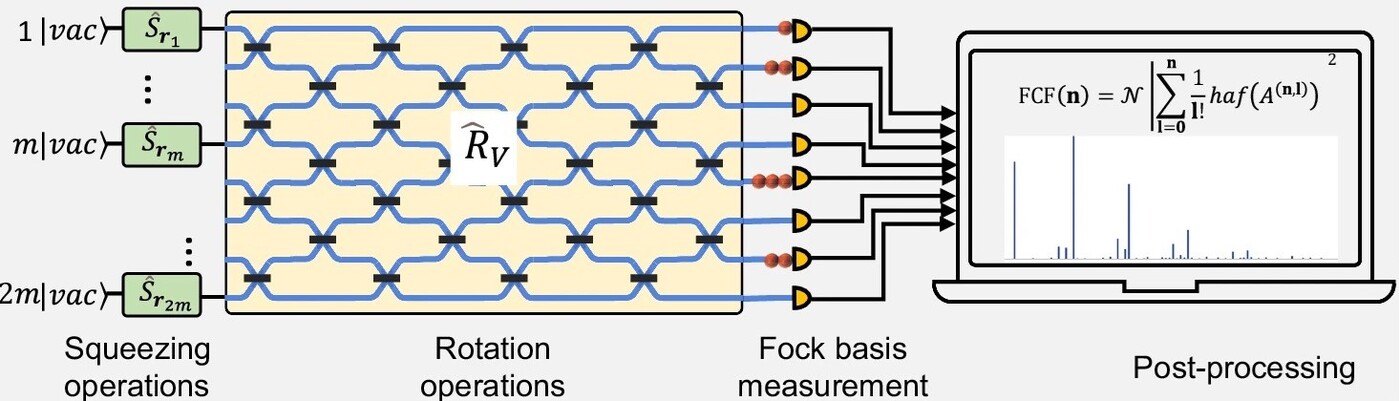
Figure 1. A squeezed vacuum state and a linear photonic network for molecular vibronic spectroscopy
The core innovation lies in employing squeezed vacuum states (quantum states with zero displacement) rather than the more experimentally demanding squeezed coherent states. These squeezed vacuum states are injected into a fully programmable linear optical network (Figure 1), enabling efficient calculation of Franck-Condon factors (FCFs) and, by extension, the full vibronic spectra of complex molecules. The photonic chip, fabricated on a silicon-on-insulator platform, integrates sixteen single-mode squeezed vacuum sources and a reconfigurable interferometer network, providing a versatile and scalable platform for quantum molecular simulation.
The algorithm is underpinned by the mapping of molecular vibrational transitions onto photon transitions in a linear optical network. The vibronic spectral profile, or Franck-Condon profile (FCP), is determined by the FCFs, which are the squared overlap integrals of vibrational wavefunctions in different electronic states. Classically, calculating FCFs for large molecules is equivalent to evaluating the loop hafnian of a matrix, which is a computationally intractable problem. The quantum approach leverages the boson sampling protocol, which links the hafnian of a matrix to the output statistics of photons in a linear optical network. By using squeezed vacuum states and appropriate mode expansion, the original displacement parameters of molecular vibrational modes can be encoded as covariances between optical modes. This allows the simulation of an m-mode molecular transition using squeezing and rotation operations on 2m optical modes.
The central equation connecting the FCF to the quantum optical experiment is:
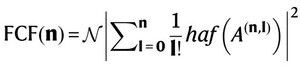
where N is a normalisation constant, haf denotes the hafnian function and A(n,l) is a submatrix determined by the output photon pattern. This equation encapsulates the quantum advantage: by sampling the output photon patterns, the experimentally approximated FCFs can be reconstructed, enabling the calculation of the full vibronic spectrum.

Figure 2. Schematic of a quantum microprocessor chip and experimental setup
a) Setup to generate single-mode squeezing
b) Photograph of the microprocessor chip
c) Overview of off-chip control and measurement devices
The integrated quantum photonic microprocessor chip realises this algorithm in hardware. The schematic of the quantum microprocessor chip and experimental setup is illustrated in Figure 2. The chip, measuring 13 × 4 mm2, incorporates sixteen nonlinear photon sources, 272 thermo-optic phase shifters, 319 multimode interferometer beam splitters and 65 optical couplers, all monolithically integrated on a silicon-on-insulator substrate. The squeezed vacuum states are generated via degenerate spontaneous four-wave mixing in spiral waveguides, while the programmable interferometer network implements arbitrary unitary transformations. Photon number-resolving detection is achieved through a pseudo-number-resolving scheme, utilising on-chip beam splitters and superconducting single-photon detectors, which enables up to four-photon resolution per mode. The entire system is controlled and stabilised via a combination of thermoelectric and water cooling, ensuring high fidelity and low-noise operation.
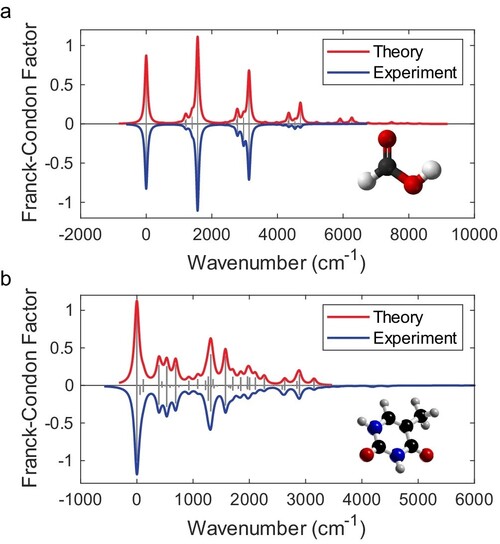
Figure 3. Vibronic spectra reconstruction of (a) formic acid (CH2O2) and (b) thymine (C5H6N2O2). The theoretical and experimental FCPs are presented as vertical grey bars above and below the x-axis, respectively. The red and blue curves are the Lorentzian broadening of the bars.
Experimental results demonstrate the power and versatility of this approach. The chip was used to simulate the vibronic spectra of formic acid (CH2O2) and thymine (C5H6N2O2) under the Condon approximation (Figure 3). For formic acid, four vibrational modes were selected and encoded into an eight-mode optical network, achieving a reconstructed fidelity of 92.9% compared to theoretical predictions. For thymine, seven modes were encoded, yielding a fidelity of 97.4%. These high fidelities are limited primarily by photon loss, detector efficiency and the maximum achievable squeezing level. Notably, the quantum approach outperformed classical strategies, with a fidelity improvement (quantum-classical gap) of 6.8% for formic acid and 7.3% for thymine at the experimental squeezing level.
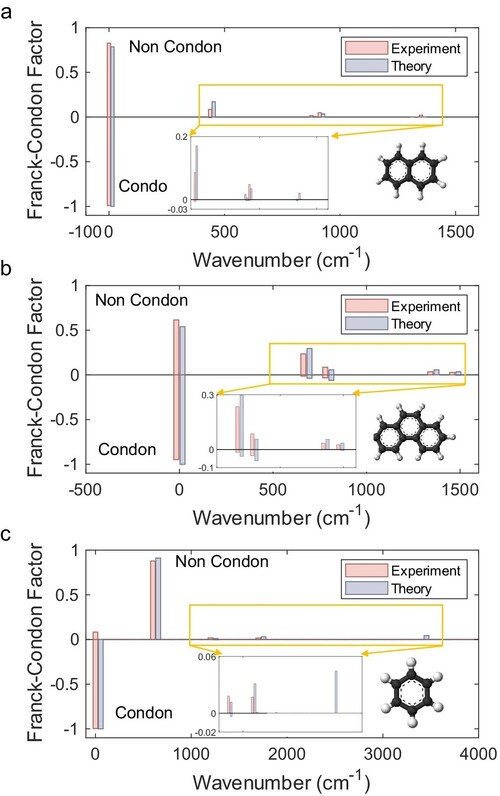
Figure 4. Vibronic spectra of (a) naphthalene (C10H8), (b) phenanthrene (C14H10) and (c) benzene (C6H6). Red graph bars show the histogram of experimental energies, while the blue bars show the theoretical results.
Beyond the Condon approximation, the chip was employed to simulate non-Condon effects, where the transition dipole moment varies with nuclear coordinates, in naphthalene (C10H8), phenanthrene (C14H10) and benzene (C6H6) (Figure 4). Using the first-order Herzberg-Teller expansion, the experimental spectra for these molecules achieved fidelities of 98.4%, 98.4% and 93.4% respectively, closely matching theoretical predictions and capturing subtle spectral features arising from non-Condon effects. These results highlight the chip’s capability to simulate complex molecular processes beyond the reach of classical computation and previous quantum experiments, which were limited to virtual or two-mode molecules.
The scalability of the approach is underpinned by rigorous analysis. The number of samples required to estimate the FCP at a desired precision ϵ scales as O(ϵ−2), representing a polynomial overhead. Importantly, the number of terms in the hafnian summation does not increase with the number of vibrational modes, ensuring that the algorithm remains efficient as system size grows. The chip’s architecture features a high degree of programmability and integration. This supports the simulation of molecules with up to seven modes, surpassing previous demonstrations. As photonic technology advances, this paves the way for even larger systems.
In conclusion, this work marks a significant advance in quantum molecular simulation, demonstrating for the first time the experimental generation of molecular vibronic spectra for real molecules with multiple modes on an integrated photonic chip. By leveraging squeezed vacuum states and a scalable linear optical network, the approach overcomes key experimental bottlenecks and achieves high-fidelity simulation of both Condon and non-Condon molecular processes. The results not only validate the theoretical framework but also establish a practical pathway towards quantum-enhanced molecular spectroscopy, with potential applications in quantum chemistry, drug discovery and quantum machine learning. As integrated photonic technology continues to mature, offering lower losses, higher squeezing and improved detection, the prospect of quantum advantage in molecular simulation draws ever closer, heralding a new era for quantum-enabled chemical and biological research.
Prof. Chin is an expert in microelectronics and bioelectronics. He has applied silicon microelectronics and nanofabrication for integrated photonic devices innovations, and employed bioelectronic chip technology with silicon semiconductors for quantum and biomedical applications. Prof. Chin has published over 70 papers in top-tier journals, including Advanced Materials, Science Translational Medicine, Advanced Science, Nature Communications, ACS Nano and npj Quantum Information.
| References |
|---|
[1] Zhu, H.H., Chen, H. S., Chen, T., Li, Y., Luo, S. B., Karim, M. F., Luo, X. S., Gao, F., Li, Q., Cai, H., Chin, L. K., Kwek, L. C., Norden, B., Zhang, X. D. & Liu, A. Q. Large-scale photonic network with squeezed vacuum states for molecular vibronic spectroscopy. Nature Communications 15, 6057 (2024). https://doi.org/10.1038/s41467-024-50060-2
 | Prof. Lip Ket CHIN |




