
So you just did a logistic regression or a nice glmer, and
you got a significant effect! Hooray! And that effect is 5. But now you're wondering... 5 whats? Is the difference between your conditions
5%, or 5 of something else? Now you're stuck staring at your model summary asking, "What does this mean?"
This post will describe what logistic regression coefficients mean, and review some quick-and-dirty (and some not-so-quick-but-still-dirty)
ways to interpret them.
While we often think of binary outcomes in terms of proportions (e.g., "92% correct responses in this condition"), logistic regression is actually based on [log] odds (which can be calculated from a proportion using the logit function). Odds can be understood as the proportion of some target outcome (e.g., correct responses) expressed in terms of how many non-target outcomes there are per target outcome. For example, a proportion of 50% (e.g., 50% accuracy) corresponds to an odds ratio of 1/1, because 50% accuracy means there is 1 incorrect response for each correct response. A proportion of 33%, on the other hand, corresponds to an odds ratio of 1/2 (2 incorrect responses for every 1 correct response), a proportion of 75% corresponds to an odds ratio of 3/1, etc. More generally, for any probability p [between 0 and 1], the odds are \(\frac{p}{1-p}\). In turn, logistic regression uses the log of the odds ratio (i.e., the logit), rather than the odds ratio itself; therefore, 50% accuracy corresponds to \(log(\frac{.5}{1-.5})=log(1/1)=0\), 33% accuracy to \(log(\frac{.33}{1-.33})=log(1/2)=-0.69\), 75% to \(log(\frac{.75}{1-.75})=log(3/1)=1.09\), etc.
Likewise, a log odds value \(\hat Y\) can be converted back into proportions using the inverse logit formula, \(\frac{e^\hat Y}{1+e^\hat Y}\). Thus, a log odds value of 0 corresponds to 50% probability (\(\frac{e^0}{1+e^0}=\frac{1}{1+1}=1/2\)), a log odds value of 2 corresponds to 88% probability (\(\frac{e^2}{1+e^2}\)), etc.
Why is this useful? Because logistic regression coefficients (e.g., in the confusing model summary from your logistic regression analysis) are reported as log odds. Log odds are difficult to interpret on their own, but they can be translated using the formulae described above. Log odds could be converted to normal odds using the exponential function, e.g., a logistic regression intercept of 2 corresponds to odds of \(e^2=7.39\), meaning that the target outcome (e.g., a correct response) was about 7 times more likely than the non-target outcome (e.g., an incorrect response). While logistic regression coefficients are sometimes reported this way, especially in the news or pop science coverage (e.g., those headlines like "bacon eaters 3.5 times more likely to comment on Youtube videos!"), I find this difficult to interpret and I prefer to think about the results in terms of proportions. Fortunately, the log odds can be turned into a proportion using the inverse logit function, as shown above.
The coefficient for an intercept is relative to 0 and thus can be straightforwardly interpreted through the inverse logit function. For example, in a design with several categorical conditions that are dummy-coded, the intercept corresponds to the predicted outcome for the reference (baseline) condition, and inverse-logit-transforming the coefficient will show the response proportion (e.g., percent accuracy) for that condition, as shown in the example below (using R code). Note that the inverse logit of the intercept is exactly the same as the raw percent of admissions from the data, and that the significant p-value for the intercept tells us that the log odds of admission are significantly less than 0, i.e., the percentage chance of admission is significantly less than 50%:
# Load some sample data admission <- read.csv("https://stats.idre.ucla.edu/stat/data/binary.csv") # Make sure 'admit' is a factor. Reference level is 'no', target level is 'yes' admission$admit <- factor( ifelse( admission$admit==1, "yes", "no" ) ) # Turn 'rank' into an unordered factor. This isn't realistic (it should be ordered) but # I just want an example for dummy coding admission$rank <- factor( admission$rank, levels=4:1 ) # Show the proportions admitted for each rank xtabs(~rank+admit, admission)[,2] / xtabs(~rank, admission)
## rank ## 4 3 2 1 ## 0.1791045 0.2314050 0.3576159 0.5409836
# Make a model summary( model1 <- glm( admit ~ rank, admission, family="binomial" ) )$coefficients
## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -1.5224265 0.3186144 -4.7782726 1.768076e-06 ## rank3 0.3220316 0.3846844 0.8371319 4.025184e-01 ## rank2 0.9366996 0.3610304 2.5945174 9.472383e-03 ## rank1 1.6867296 0.4093073 4.1209370 3.773346e-05
# Inverse logit transform the model coefficients into proportions, to show that the # log odds of the intercept corresponds to its proportion but none of the other # model coefficients do exp( coef(model1) ) / ( 1 + exp( coef(model1) ) )
## (Intercept) rank3 rank2 rank1 ## 0.1791045 0.5798193 0.7184325 0.8437936
However, interpreting other model coefficients is not as straightforward. Look at the example above: the admission rate for rank "3" was 23.1%, which is 5.2 percentage points greater than the baseline admission rate of 17.9% for rank "4". In the model, this is represented by a coefficient of 0.3220316, indicating this much greater log odds of acceptance in this condition, compared to the intercept (rank "4"). However, simply transforming that coefficient with the inverse logit function yields a value that corresponds neither to the actual percent admission in that condition, nor the difference in percent admission between that and the baseline. This is because the inverse function assumes the log odds are relative to 0, whereas the regression coefficient for this condition is relative to the intercept (-1.5). To get a meaningful proportion, we have to instead run the logit function on the predicted value for this condition (i.e., plugging the coefficients into the regression equation), as shown below:
# The predicted value for this condition: the intercept, plus the dummy coefficient for this condition Yhat_rank3 <- coef(model1)["(Intercept)"] + coef(model1)["rank3"] # Converting the predicted value from log odds into percentage exp( Yhat_rank3 ) / ( 1 + exp( Yhat_rank3 ) )
## (Intercept) ## 0.231405
Notice that now we get a value which corresponds exactly to the percentage admittance for that condition. This is pretty uninteresting, since it tells us what we already saw in the data, but it is a nice demonstration of the relationship between log odds and proportions. (Plus, the non-significant coefficient for "rank3" in the model tells us that this proportion of 23.1% is not significantly higher than the 17.9% acceptance rate for rank 4, i.e., the intercept.) These estimates can, however, give more insight in cases where the data are noisier: for example, if you have repeated measures for subjects and items and thus used a mixed model (in which case model predictions won't exactly match the data, and thus the model predictions converted into percentages are useful for illustrating the data pattern once subject and item variance has been accounted for) or if the predictor (independent variable) is not categorical but continuous (in which case the predicted proportion won't correspond to the percent admittance for any one condition, but to the slope of the regression line). Let's look at an example with a continuous predictor.
# A logistic model: probability of being admitted, as a function of GPA (grade point average) summary( model2 <- glm( admit ~ gpa, admission, family="binomial" ) )$coefficients
## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -4.357587 1.0353170 -4.20894 2.565715e-05 ## gpa 1.051109 0.2988694 3.51695 4.365354e-04
### Plot the data and model # Plot the actual datapoints, with a little bit of jitter plot( admission$gpa, jitter(ifelse( admission$admit=="yes",1,0 ),.1), pch=20, xlab="GPA", ylab="Likelihood of admittance" ) # Inverse logit function inverselogit <- function(x){ exp(x) / (1+exp(x) ) } # Get the unique gpas gpas <- sort( unique( admission$gpa ) ) # Plot the model prediction for each GPA, with a line through them lines( gpas, inverselogit( coef(model2)["(Intercept)"] + gpas*coef(model2)["gpa"] ), type="o", pch=22, bg="blue", cex=2 )
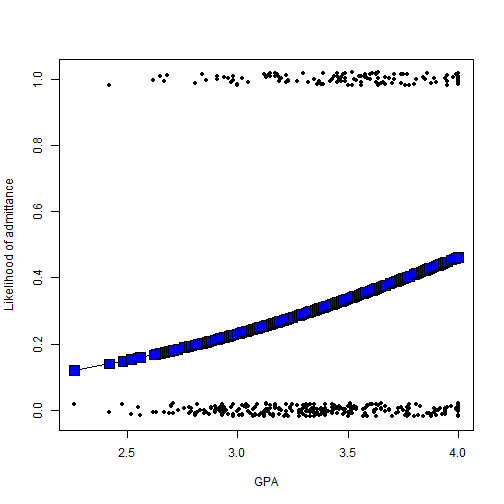
Notice here that converting the model predictions into proportions, using the inverse logit function, helps us interpret the result: we know that the positive model coefficient for "gpa" should indicate that likelihood of being admitted increases as GPA increases, and the regression line with blue squares in the figure indeed illustrates this pattern.
However, there is a new challenge now. With linear OLS regression, model coefficients have a straightforward interpretation: a model coefficient b means that for every one-unit increase in \(x\), the model predicts a b-unit increase in \(\hat Y\) (the predicted value of the outcome variable). E.g., if we were using GPA to predict test scores, a coefficient of 10 for GPA would mean that for every one-point increase in GPA we expect a 10-point increase on the test. Technically, the logistic regression coefficient means the same thing: as GPA goes up by 1, the log odds of being accepted go up by 1.051109. However, log odds are difficult to interpret, especially relative log odds: while we can straighforwardly calculate that a log odds of 1 on its own means a 73.1% chance of acceptance, an increase of 1 in log odds means something different depending on what the log odds increased from—as we can see by the fact that the line above is not quite straight. What we (or at least I) would really like to know is how much the percentage likelihood of being accepted goes up, on average, for each one-unit increase in GPA. To do this, we need to calculate the marginal effects.
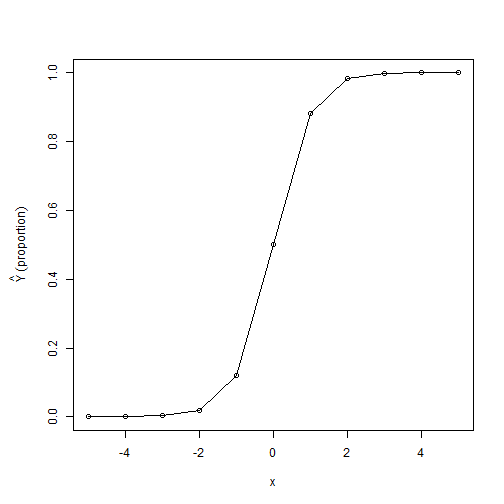
When you say how much of an increase there is in \(\hat Y\) for every one-unit increase in \(x\), you are describing the marginal effect. (This is not to be confused with the other sense in which we might use the phrase "marginal effect", to describe an effect that is not quite statistically significant. Rather, this term is like the "marginal means" we might look at in an ANOVA design.) As we noted above, linear regression coefficients directly correspond to marginal effects: if we regress test score on GPA and find a coefficient of 10, that means that a 1-point increase in GPA corresponds to a predicted 10-point increase in test score. Logistic regression coefficients also correspond to marginal effects, but the unit of measurement is not test points or whatever; instead, the unit of measurement is log odds, and and a 1-point increase in log odds is difficult to put in context. We know how to transform log odds into proportions using the inverse logit function described above, but since this is a non-linear transform, expressing the marginal effect in terms of proportions rather than log odds is not straightforward. This is because the logistic regression line is not straight, and thus the marginal effect expressed as a proportion is not stable across the range of data. We can illustrate this with the plot below, illustrating a fake logistic regression with an intercept of 0 and a coefficient of 2: notice that stepping from \(x=0\) to \(x=1\) brings along a large increase in the predicted proportion (about a 40% increase), whereas stepping from \(x=4\) to \(x=5\) brings along almost no increase, as the predicted probability was already near ceiling.
par(mar=c(5.1,5.1,4.1,2.1) ) plot( -5:5, inverselogit(2*(-5:5)), type="o", xlab="x", ylab=expression( hat(Y)~(proportion) ) ) par(mar=c(5.1,4.1,4.1,2.1) )
Several solutions have been proposed for this problem:
The divide-by-4 rule (see, e.g., here and here), apparently based on Gelman & Hill (2007), is the simplest to apply: it just amounts to dividing the logistic regression coefficient by 4 to get a quick-and-dirty estimate of its marginal effect. This is, of course, an oversimplification, and it has limitations (e.g., per Ben Bolker's comment at the linked blog post, it is mainly just reasonable for coefficients with a small absolute value, and apparently Gelman & Hill (2007) discuss its limitations in more detail).
The marginal-effect-at-mean method, based on Kleiber & Zeileis (2008), is described here. Getting the marginal effect for the mean value of \(x\) amounts to calculating the predicted proportion when all the predictor variables are held to their mean, and then also calculating the predicted proportion when all the other predictor variables are at their mean and the variable of interest (i.e., the one for whose coefficient we are trying to find the marginal effect) is at mean+1, and then taking the difference of these two predicted proportions. (A variation of this method is to calculate the derivative of the regression line at this particular spot [see here]; recall from intro calculus that the derivative is the slope of a curvy line at one particular infinitesimal point along the line.)
The average-of-marginal-effects method, also from Kleiber & Zeileis (2008) and described here, amounts to taking the mean of all predicted values (technically these are not the predicted proportions, but the probability density function of all the predicted log-odds values) and multiplying by the logistic regression coefficient.
Because I am not particularly familiar with any of these and don't know mathematically what the pros and cons of each are, let's try calculating all three and seeing how they line up with the actual predicted values from our data. The chunk of code below first calculates the three kinds of marginal effects, and then plots the data in several ways so we can compare the marginal effects against what we see.
# Grab the model coefficients intercept <- coef( model2 )["(Intercept)"] gpacoef <- coef( model2 )["gpa"] ### Marginal effect using the divide-by-4 rule div4marginal <- gpacoef/4 ### Done getting divide-by-4 marginal effect ### Marginal effect at the mean value of GPA # First get the mean value of GPA. Note that here I'm taking the mean of all the gpa values, not # the mean of the unique values [i.e., not the exact middle of the range of gpa values, which would # be given by mean( range( admission$gpa ) )]. I'm not sure, however, which would be better. meangpa <- mean( admission$gpa ) # The predicted value at this spot + 1, minus the predicted value at this spot marginal_effect_at_mean <- inverselogit( intercept + gpacoef*(meangpa+1) ) - inverselogit( intercept + gpacoef*meangpa ) ### Done getting marginal effect at mean ### Mean of marginal effects # The mean of the PDF of the predicted values [in log odds], times the coefficient. # www.ucd.ie/t4cms/WP11_22.pdf#page=5 has a function that does this easily, and can also # give standard errors of the marginal effects mean_of_sample_marginal_effects <- mean( dlogis( intercept+gpacoef*admission$gpa ) )*gpacoef ### # Show the different marginal effects on the screen ( marginal <- lapply( list( div_by_4=div4marginal, at_mean=marginal_effect_at_mean, mean_of_marginal=mean_of_sample_marginal_effects ), as.numeric ) )
## $div_by_4 ## [1] 0.2627772 ## ## $at_mean ## [1] 0.2526013 ## ## $mean_of_marginal ## [1] 0.2204751
### Create plots showing the marginal effects over the range of the data par( mfrow=c(1,3), mar=c(5.6, 4.1, 4.1, 2.1) ) # The range of GPAs in the data gpas <- seq( min(admission$gpa), max(admission$gpa), by=.1 ) # The predicted likelihood of acceptance for each GPA proportion_accepted <- inverselogit( intercept + gpacoef*gpas ) # Plot the predicted proportion accepted over the whole range of data; i.e., the # same regression line we saw above plot( gpas, proportion_accepted, xlab="GPA", ylab="Proportion accepted", pch=16, cex=3, main="Predicted outcome", type="o" ) # Next we'll plot the change in likelihood of acceptance, at each level of GPA. We'll do # this by comparingthe likelihood of at acceptance between GPAs of 2.96 and 2.96 (for # example), 2.5 and 3.5, etc. etc. # Here we calculate the differences. Since the GPAs are in steps of .1 (in the sequence we made # above), we know that 3.5 is 10 above 2.5 (for example), so we make two vectors which are # offset by 10 highergpas <- 11:length(proportion_accepted) lowergpas <- 1:length( proportion_accepted[-(1:10)] ) # Then we subtract the lower ones from the higher ones changes <- c( rep(NA,10), proportion_accepted[highergpas] - proportion_accepted[lowergpas] ) # Those same changes, but now instead of representing them as raw percentages (i.e., going from 15% up to # 32% would be called a 17-percentage-point increase), we now represent them as a change relative to # the starting point (i.e., going from 15% up to 32% is a 17/15 = 113% increase) changes_as_proportion <- c( rep(NA,10), (proportion_accepted[highergpas] - proportion_accepted[lowergpas])/proportion_accepted[lowergpas] ) # Plot the raw changes plot( gpas, changes, xlab="GPA", ylab="Change in proportion accepted for a one-unit increase in frequency", xaxt="n", pch=16, cex=3, main="Marginal changes" ); axis(1, at=gpas[highergpas], labels=paste( gpas[lowergpas], "to", gpas[highergpas]), las=2 ) # Plot the proportional changes plot( gpas, changes_as_proportion, xlab="GPA", ylab="Change in proportion accepted, as proportion", xaxt="n", pch=16, cex=3, main="Relative marginal changes" ); axis(1, at=gpas[highergpas], labels=paste( gpas[lowergpas], "to", gpas[highergpas]), las=2 )
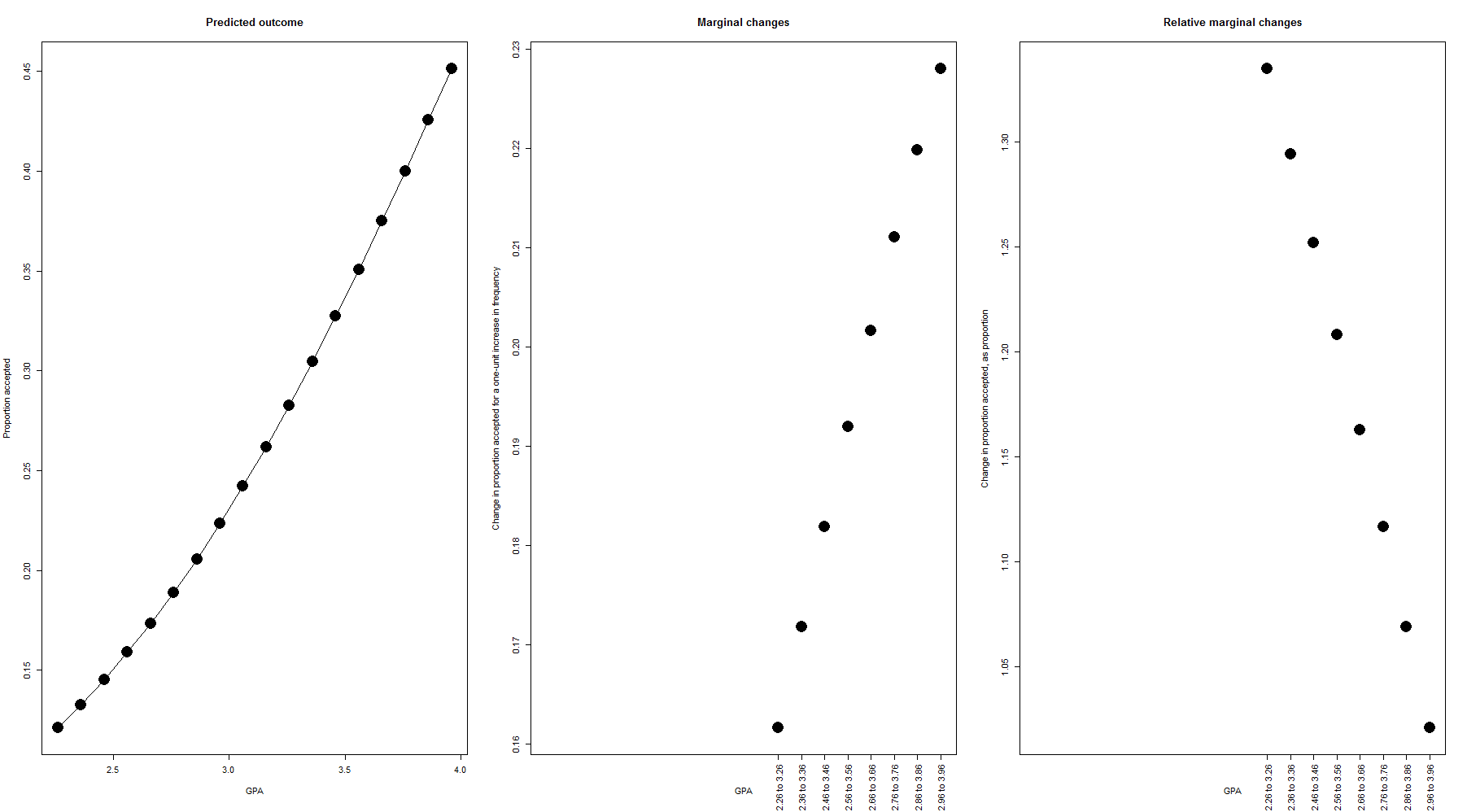
We see that the divide-by-4 rule estimated a marginal effect of 26%, the marginal-effect-at-mean-x rule estimated a marginal effect of 25%, and the mean-of-marginal-effects rule estimated a marginal effect of 22%. How do these measure up to the actual predictions? From the left-hand portion of the plot, we can see (as we saw above) that the effect of GPA is not only positive, but increasing. Over the range of GPAs tested, stepping from a low GPA to a slightly higher, but still low, GPA, confers a slightly smaller increase in acceptance likelihood than stepping from an already-high GPA to an even higher one. We can see this even more directly in the middle portion of the plot, which shows that stepping from 2.26 to 3.26 GPA is associated with a 16 percentage-point increase in likelihood of acceptance, whereas stepping from a 2.36 to a 3.36 is associated with a 17 percentage-point increase, etc. (This effect is, of course, limited to the rather narrow range of GPAs shown here; since proportions are bounded between 0% and 100%, eventually the predictions would level out. Technically speaking, this means that while the first derivative of the regression line [the marginal effect] will remain positive, the second derivative [whether the marginal effect is getting bigger and bigger or smaller and smaller] will become negative as the line asymptotes. In this dataset we just do not observe that since GPA is never greater than 4.)
To me, all of the marginal estimates look like slight overestimates of the pattern we see in the middle portion of the figure, but the mean-of-marginal-effects method gives the least serious overestimate. Of course, in a way, every marginal estimate will be "wrong", in that a single number will not capture the marginal effect at every point along the range of x-values (as discussed above); in this case, the mean-of-marginal-effects method just happened to look least wrong for this particular spot in the range of x-values. I suspect that the other two measures may have been more influenced by higher potential values of GPA: for example, a change in GPA from 3.5 to 4.5 would indeed be associated with a 26 percentage-point increase, we just do not see this since the maximum value of GPA is 4 (in the United States educational system). Therefore, it would also be worthwhile to try this out with an example that has a wider range of x-values.
Let's try another such example. This one uses a different dataset, with a slightly wider range of values for the predictor.
Furthermore, this will use a mixed-effects logistic model (glmer()) rather than a standard logistic regression. This raises
some extra interesting questions, which we'll discuss below. First, let's run the code to see the model coefficients, the marginal
estimates, and the plots:
# Load libraries library(lme4) library(languageR) # A model regressing the likelihood of error (the baseline level of Correct is "incorrect") # on word frequency summary( model3 <- glmer( Correct ~ Frequency + (Frequency|Subject) + (1|Word), lexdec, family="binomial" ) )$coefficients
## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -1.7885391 0.9586225 -1.865739 0.06207794 ## Frequency -0.5173847 0.2069102 -2.500528 0.01240084
# Grab the model coefficients intercept <- fixef( model3 )["(Intercept)"] freqcoef <- fixef( model3 )["Frequency"] ### Marginal effect using the divide-by-4 rule div4marginal <- freqcoef/4 ### Done getting divide-by-4 marginal effect ### Marginal effect at the mean value of Frequency # First get the mean value of Frequency meanfreq <- mean( lexdec$Frequency ) # The predicted value at this spot + 1, minus the predicted value at this spot marginal_effect_at_mean <- inverselogit( intercept + freqcoef*(meanfreq+1) ) - inverselogit( intercept + freqcoef*meanfreq ) ### Done getting marginal effect at mean ### Mean of marginal effects # The mean of the PDF of the predicted values [in log odds] based only on fixed effects, times the coefficient. mean_of_sample_marginal_effects <- mean( dlogis( intercept+freqcoef*lexdec$Frequency ) )*freqcoef ### ### Mean of marginal effects # The mean of the PDF of the predicted values [in log odds] based on both fixed and randome ffects, times the coefficient. # www.ucd.ie/t4cms/WP11_22.pdf#page=7 has a function that does this easily, and can also # give standard errors of the marginal effects mean_of_sample_marginal_effects_fitted <- mean( dlogis( log( fitted(model3)/(1-fitted(model3)) ) ) )*freqcoef ### # Show the different marginal effects on the screen ( marginal <- lapply( list( div_by_4=div4marginal, at_mean=marginal_effect_at_mean, mean_of_marginal=mean_of_sample_marginal_effects, mean_of_marginal_fitted=mean_of_sample_marginal_effects_fitted ), as.numeric ) )
## $div_by_4 ## [1] -0.1293462 ## ## $at_mean ## [1] -0.005650974 ## ## $mean_of_marginal ## [1] -0.008729906 ## ## $mean_of_marginal_fitted ## [1] -0.01583169
### Create plots showing the marginal effects over the range of the data par( mfrow=c(1,3), mar=c(5.6, 4.1, 4.1, 2.1) ) # The range of frequenciess in the data freqs <- seq( min(lexdec$Frequency), max(lexdec$Frequency), by=.1 ) # The predicted likelihood of acceptance for each frequency proportion_error <- inverselogit( intercept + freqcoef*freqs ) # Plot the predicted proportion accepted over the whole range of data; i.e., the # same regression line we saw above plot( freqs, proportion_error, xlab="Freq", ylab="Predicted roportion of errors", pch=16, cex=3, main="Predicted outcome", type="o" ) # Next we'll plot the change in likelihood of acceptance, at each level of frequency. We'll do # this by comparing the likelihood of at acceptance between freqss of 2.96 and 2.96 (for # example), 2.5 and 3.5, etc. etc. # Here we calculate the differences. Since the frequenciess are in steps of .1 (in the sequence we made # above), we know that 3.5 is 10 above 2.5 (for example), so we make two vectors which are # offset by 10 higherfreqs <- 11:length(proportion_error) lowerfreqs <- 1:length( proportion_error[-(1:10)] ) # Then we subtract the lower ones from the higher ones changes <- c( rep(NA,10), proportion_error[higherfreqs] - proportion_error[lowerfreqs] ) # Those same changes, but now instead of representing them as raw percentages (i.e., going from 15% up to # 32% would be called a 17-percentage-point increase), we now represent them as a change relative to # the starting point (i.e., going from 15% up to 32% is a 17/15 = 113% increase) changes_as_proportion <- c( rep(NA,10), (proportion_error[higherfreqs] - proportion_error[lowerfreqs])/proportion_error[lowerfreqs] ) # Plot the raw changes plot( freqs, changes, xlab="", ylab="Change in proportion error for a one-unit increase in frequency", xaxt="n", pch=16, cex=3, main="Marginal changes" ) axisfreqs <- round( seq( 1, length(higherfreqs), length.out=length(higherfreqs)/3 ) ) axis(1, at=freqs[higherfreqs[axisfreqs]], labels=paste( round( freqs[lowerfreqs[axisfreqs]], 1), "to", round( freqs[higherfreqs[axisfreqs]], 1) ), las=2 ) # Plot the proportional changes plot( freqs, changes_as_proportion, xlab="", ylab="Change in proportion error, as proportion", xaxt="n", pch=16, cex=3, main="Relative marginal changes" ) axis(1, at=freqs[higherfreqs[axisfreqs]], labels=paste( round( freqs[lowerfreqs[axisfreqs]], 1), "to", round( freqs[higherfreqs[axisfreqs]], 1) ), las=2 )
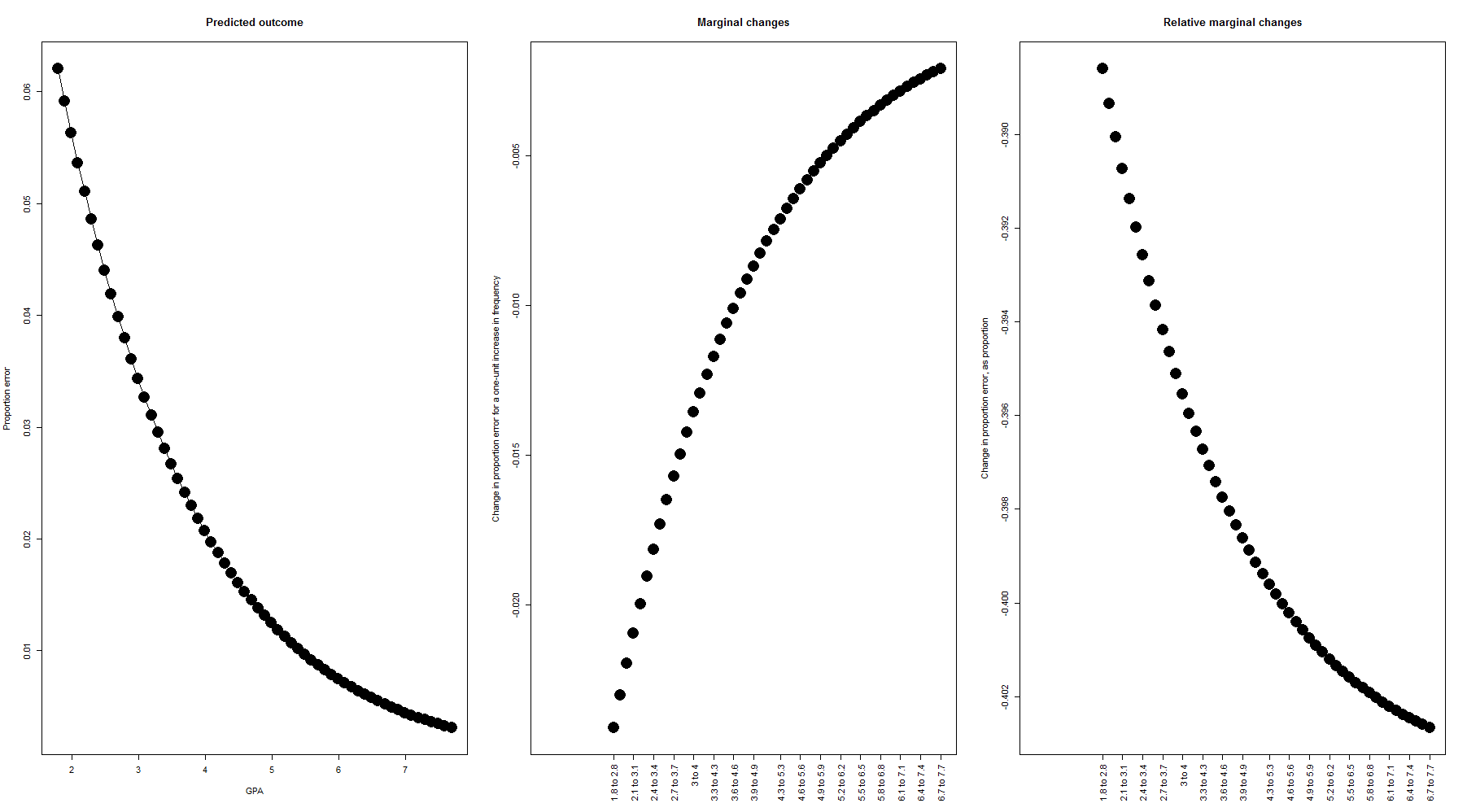
On the left-hand portion of the figure we see that, while the likelihood of making an incorrect response is always low even at low frequencies (participants were very accurate), it gets even lower as frequency increases. The center portion shows us that the magnitude of the marginal effect decreases as frequency increases. How did our marginal effect estimates measure up against this plot?
The marginal effect estimated with the divide-by-4 rule is clearly way off: it estimates a much lower marginal effect (i.e., much bigger decrease in the probability of an incorrect response) than we ever actually observe. I suspect this is because the model is already at such extreme negative log odds (i.e., it's already making predictions that are all close to 0% probability); if we had much lower x-values (such as negative frequencies, which are of course linguistically impossible, but which the model still fits) then the divide-by-4 estimate of the marginal effect would be more accurate in that range. In other words, as warned in one of the blog posts linked above, this estimate is probably only meaningful when the actual outcomes are in more middling probability ranges, and not so meaningful when we're dealing with very high or very low target response proportions.
The marginal effect at the mean frequency, on the other hand, appears reasonable, as does the mean of the marginal effects. Both correspond to effects near the middle of the frequency distribution, as expected. You will also note that I calculated the mean-of-marginal-effects in two different ways. The first used the predicted values based only on the fixed effects, whereas the second used the fitted values, i.e., the predicted values based on both the fixed effects and the random effects. The latter is what is implemented in Alan Fernihough's function. In this case, however, the former gave what looks to me like a more reasonable estimate, and conceptually speaking it also seems more relevant, for me, since I usually only care about the fixed effects. Nonetheless, probably either one could be appropriate.
Keep in mind that there is actually no single "right" estimate of the marginal effect, when we're expressing it as a proportion/percentage. This is because, as noted several times above, the regression line is not straight: the marginal effect can be quite different depending on where along the x-axis you look. Therefore, I cannot claim that any one of these measures is unequivocally the best. Probably each one has its biases, and I haven't done enough simulations or math to fully understand what they all are—although, as we observed above, it seems like the divide-by-4 method only gives a good estimate of the marginal effect near whatever x-values are yielding middling predicted proportions, and not in the ranges where the predicted proportions are very extreme. From the datasets I have tried this on, such as the ones we see above, the mean of the marginal effects based on fixed-effect predictors seems like an ideal estimate. Nevertheless, as we have seen above, none of these estimates is perfect, and none is a substitute for plotting the predictions. (Also note that marginal effect estimates like these are unnecessary in models with only categorical predictors, where the exact marginal effects can be calculated by simply comparing two conditions or combinations of conditions.)
by Stephen Politzer-Ahles. Last modified on 2016-05-30.